Gromacs学习
cmake ../gromacs-2022.1/ \-DFFTW_INCLUDE_DIR=$FFTW_LOCATION/include \-DFFTW3F_LIBRARIES=$FFTW_LOCATION/lib/libfftw3.a \-DCMAKE_INSTALL_PREFIX=$(pwd) \-DGMX_Xll=OFF \-DCMAKE_CXX_COMPILER=$MPICCDIR/mpic++ \-DCMAKE_C_COMPILER=$MPICCDIR/mpicc \-DGMX_MPI=ON \-DGMX_PREFER_STATIC_LIBS=ON

1.准备所需的pdb格式文件 gmx pdb2gmx (从库里下载的pdb文件转成gromacs的坐标):-f(输入pdb文件)-o(输出gro文件)-p(输出top文件)-i(输出itp文件)-n(输出ndx)-ignh(舍弃H,系统会自动加上)-ter(肽链两端质子化戴帽子)“-water”指定使用的水模型
gmx pdb2gmx -f 1AKI_clean.pdb -o 1AKI_processed.gro -water spce -ignh
2.检查拓扑文件 3.定义单位盒子并填充溶剂 定义盒子: gmx editconf的主要功能是对体系结构进行编辑, 也可以将通用结构格式保存或转换为.gro, .g96或.pdb等其他格式. 选项-bt设定盒子类型, triclinic为三斜盒子, cubic为所有边长都相等的长方体盒子(即立方体盒子), dodecahedron代表菱形十二面体盒子(等边十二面体), octahedron为截角八面体盒子(即将两个底面重合的四面体切去方向相反的两头, 同时保证所有的边长相等). 后面两种盒子是三斜盒子的特殊情况. 截角八面体三个盒向量的长度是两个相对六边形之间的最短距离. 相对于具有周期性映象距离的立方盒子, 具有相同周期距离的菱形十二面体盒子的体积是立方盒子的71%, 而截角八面体盒子的体积是立方盒子的77%. -d选项指定体系中的原子到盒子编边界的最小距离. 使用-d选项时, 对三斜盒子会使用体系在x, y和z方向的大小, 对立方盒子, 菱形十二面体盒子或截角八面体盒子, 盒子的大小被设定为体系直径(原子间的最大距离)加上两倍的指定距离. -c选项使分子在盒子中居中
gmx editconf -f 1AKI_processed.gro -o 1AKI_newbox.gro -c -d 1.0 -bt cubic
添加溶剂: gmx solvate能够完成以下两项任务:
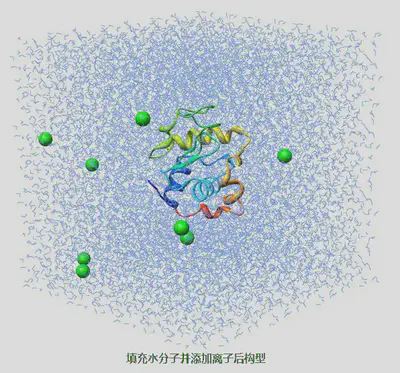
创建一个充满溶剂的盒子. 可以通过指定-cs和-box选项来完成. 对具有盒子信息但不含原子的结构文件则可以通过指定-cs和-cp来实现. 将溶质分子, 如蛋白质进行溶剂化, 使其处于溶剂分子的包围之中. -cp和-cs分别用于指定溶质和溶剂. 不设定-box时, 会使用溶质坐标文件(-cp)中的盒子信息. 如果你希望将溶质置于盒子的中心, 可以使用gmx editconf命令, 它有非常多的选项用于改变盒子的规格和使分子居中. 对某一位置, 若溶质分子中任意原子与溶剂分子中任意原子之间的距离小于这两个原子的范德华半径之和, 则会将溶剂分子从盒子中移除. 程序会读取数据文件(vdwradii.dat)中的范德华半径, 并根据-scale选项的设置进行缩放. 若不能在数据文件中找到所需的半径值, 相应的原子将通过-radius来设定(未缩放)距离.
gmx solvate -cp 1AKI_newbox.gro -cs spc216.gro -o 1AKI_solv.gro -p topol.top
4.添加离子 ions.tpr文件是结构/状态文件 ions.mdp设置分子动力学参数: integrator指定算法,steep是最速下降法。emstep是初始步长,默认为0.01。emtol指定当最大力值到1000时停止能量最小化。nsteps指定最大步数,设置为-1时没有上限。 nstlist指定更新邻居列表的频率
; ions.mdp - used as input into grompp to generate ions.tpr
; Parameters describing what to do, when to stop and what to save
integrator = steep ; Algorithm (steep = steepest descent minimization)
emtol = 1000.0 ; Stop minimization when the maximum force < 1000.0 kJ/mol/nm
emstep = 0.01 ; Energy step size
nsteps = 50000 ; Maximum number of (minimization) steps to perform
; Parameters describing how to find the neighbors of each atom and how to calculate the interactions
nstlist = 1 ; Frequency to update the neighbor list and long range forces
cutoff-scheme = Verlet
ns_type = grid ; Method to determine neighbor list (simple, grid)
coulombtype = PME ; Treatment of long range electrostatic interactions
rcoulomb = 1.0 ; Short-range electrostatic cut-off
rvdw = 1.0 ; Short-range Van der Waals cut-off
pbc = xyz ; Periodic Boundary Conditions (yes/no)
gmx grompp生成运行输入文件
gmx grompp -f ions.mdp -c 1AKI_solv.gro -p topol.top -o ions.tpr
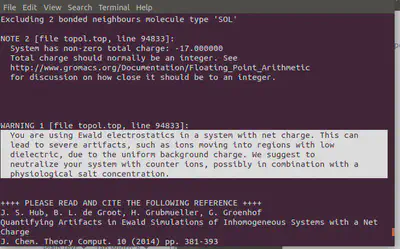
gmx genion -s ions.tpr -o 1AKI_solv_ions.gro -p topol.top -pname NA -nname CL -nn 8
; minim.mdp - used as input into grompp to generate em.tpr
integrator = steep ; Algorithm (steep = steepest descent minimization)
emtol = 1000.0 ; Stop minimization when the maximum force < 1000.0 kJ/mol/nm
emstep = 0.01 ; Energy step size
nsteps = 50000 ; Maximum number of (minimization) steps to perform
; Parameters describing how to find the neighbors of each atom and how to calculate the interactions
nstlist = 1 ; Frequency to update the neighbor list and long range forces
cutoff-scheme = Verlet
ns_type = grid ; Method to determine neighbor list (simple, grid)
coulombtype = PME ; Treatment of long range electrostatic interactions
rcoulomb = 1.0 ; Short-range electrostatic cut-off
rvdw = 1.0 ; Short-range Van der Waals cut-off
pbc = xyz ; Periodic Boundary Conditions (yes/no)
gmx grompp -f minim.mdp -c 1AKI_solv_ions.gro -p topol.top -o em.tpr
确保更新了top文件后,执行mdrun能量最小化:
gmx mdrun -v -deffnm em
em.log: ASCII文本的日志文件, 记录了能量最小化过程 em.edr: 二进制能量文件 em.trr: 全精度的二进制轨迹文件 em.gro: 能量最小化后的结构
gmx energy:将能量写入xvg文件并显示均值
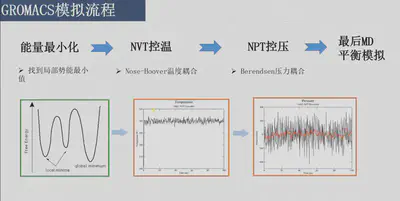
6.平衡温度NVT 该步骤用于平衡体系的温度 通常情况下, 50 ps到100 ps就足够了, 因此我们进行100 ps的NVT平衡:2*50000=100 ps nvt.mdp: constraint_algorithm = lincs指定了线性约束算法
title = OPLS Lysozyme NVT equilibration
define = -DPOSRES ; position restrain the protein
; Run parameters
integrator = md ; leap-frog integrator
nsteps = 50000 ; 2 * 50000 = 100 ps
dt = 0.002 ; 2 fs
; Output control
nstxout = 500 ; save coordinates every 1.0 ps
nstvout = 500 ; save velocities every 1.0 ps
nstenergy = 500 ; save energies every 1.0 ps
nstlog = 500 ; update log file every 1.0 ps
; Bond parameters
continuation = no ; first dynamics run
constraint_algorithm = lincs ; holonomic constraints
constraints = all-bonds ; all bonds (even heavy atom-H bonds) constrained
lincs_iter = 1 ; accuracy of LINCS
lincs_order = 4 ; also related to accuracy
; Neighborsearching
cutoff-scheme = Verlet
ns_type = grid ; search neighboring grid cells
nstlist = 10 ; 20 fs, largely irrelevant with Verlet
rcoulomb = 1.0 ; short-range electrostatic cutoff (in nm)
rvdw = 1.0 ; short-range van der Waals cutoff (in nm)
; Electrostatics
coulombtype = PME ; Particle Mesh Ewald for long-range electrostatics
pme_order = 4 ; cubic interpolation
fourierspacing = 0.16 ; grid spacing for FFT
; Temperature coupling is on
tcoupl = V-rescale ; modified Berendsen thermostat
tc-grps = Protein Non-Protein ; two coupling groups - more accurate
tau_t = 0.1 0.1 ; time constant, in ps
ref_t = 300 300 ; reference temperature, one for each group, in K
; Pressure coupling is off
pcoupl = no ; no pressure coupling in NVT
; Periodic boundary conditions
pbc = xyz ; 3-D PBC
; Dispersion correction
DispCorr = EnerPres ; account for cut-off vdW scheme
; Velocity generation
gen_vel = yes ; assign velocities from Maxwell distribution
gen_temp = 300 ; temperature for Maxwell distribution
gen_seed = -1 ; generate a random seed
gmx grompp -f nvt.mdp -c em.gro -r em.gro -p topol.top -o nvt.tpr
gmx mdrun -deffnm nvt
7.平衡压力NPT 该步骤用于稳定体系的压力 npt.mdp:
title = OPLS Lysozyme NPT equilibration
define = -DPOSRES ; position restrain the protein
; Run parameters
integrator = md ; leap-frog integrator
nsteps = 50000 ; 2 * 50000 = 100 ps
dt = 0.002 ; 2 fs
; Output control
nstxout = 500 ; save coordinates every 1.0 ps
nstvout = 500 ; save velocities every 1.0 ps
nstenergy = 500 ; save energies every 1.0 ps
nstlog = 500 ; update log file every 1.0 ps
; Bond parameters
continuation = yes ; Restarting after NVT
constraint_algorithm = lincs ; holonomic constraints
constraints = all-bonds ; all bonds (even heavy atom-H bonds) constrained
lincs_iter = 1 ; accuracy of LINCS
lincs_order = 4 ; also related to accuracy
; Neighborsearching
cutoff-scheme = Verlet
ns_type = grid ; search neighboring grid cells
nstlist = 10 ; 20 fs, largely irrelevant with Verlet scheme
rcoulomb = 1.0 ; short-range electrostatic cutoff (in nm)
rvdw = 1.0 ; short-range van der Waals cutoff (in nm)
; Electrostatics
coulombtype = PME ; Particle Mesh Ewald for long-range electrostatics
pme_order = 4 ; cubic interpolation
fourierspacing = 0.16 ; grid spacing for FFT
; Temperature coupling is on
tcoupl = V-rescale ; modified Berendsen thermostat
tc-grps = Protein Non-Protein ; two coupling groups - more accurate
tau_t = 0.1 0.1 ; time constant, in ps
ref_t = 300 300 ; reference temperature, one for each group, in K
; Pressure coupling is on
pcoupl = Parrinello-Rahman ; Pressure coupling on in NPT
pcoupltype = isotropic ; uniform scaling of box vectors
tau_p = 2.0 ; time constant, in ps
ref_p = 1.0 ; reference pressure, in bar
compressibility = 4.5e-5 ; isothermal compressibility of water, bar^-1
refcoord_scaling = com
; Periodic boundary conditions
pbc = xyz ; 3-D PBC
; Dispersion correction
DispCorr = EnerPres ; account for cut-off vdW scheme
; Velocity generation
gen_vel = no ; Velocity generation is off
gmx grompp -f npt.mdp -c nvt.gro -r nvt.gro -t nvt.cpt -p topol.top -o npt.tpr
gmx mdrun -deffnm npt
8.MD 在两个系综平衡完成后,进行MD收集数据,需要进行1ns的MD模拟